SNPs, ChIPs and RNA
Attempts to understand complex phenotypes
Peter Humburg
15th April 2015
Overview
Introduction
Interpreting genomic variation
- Sequencing of patient genomes increasingly common
- Can identify relevant variants
- … amongst a large number of unrelated variants
- Computational strategies can narrow the set of candidates
- … but variants are often difficult to interpret
- Want to leverage existing data as much as possible
Exome sequencing
Breast and Ovarian Cancer risk genes
- Several DNA repair genes implicated in breast and ovarian cancer susceptability.
- Strong evidence that rare loss-of-function variants confer increased risk.
- Sequenced exons of 507 DNA repair genes in 1,150 patients.
- Sequenced pools of 24 individuals.
- Included 79 individuals with known mutations in breast cancer predisposition genes as positive controls.
Analysis strategy
- Sequence pools with HiSeq2000
- \(\gt\) 480\(\times\) coverage in 90% of target region
- Call variants in pools with Syzygy
- Good sensitivity for rare variants
(24/26 SNPs and 51/54 indels) - Identified 34,564 variants
- Good sensitivity for rare variants
- Functional annotation obtained via EnsEMBL
- Substantial clean-up and curation of annotations
- Focused on 1,044 protein truncating variants
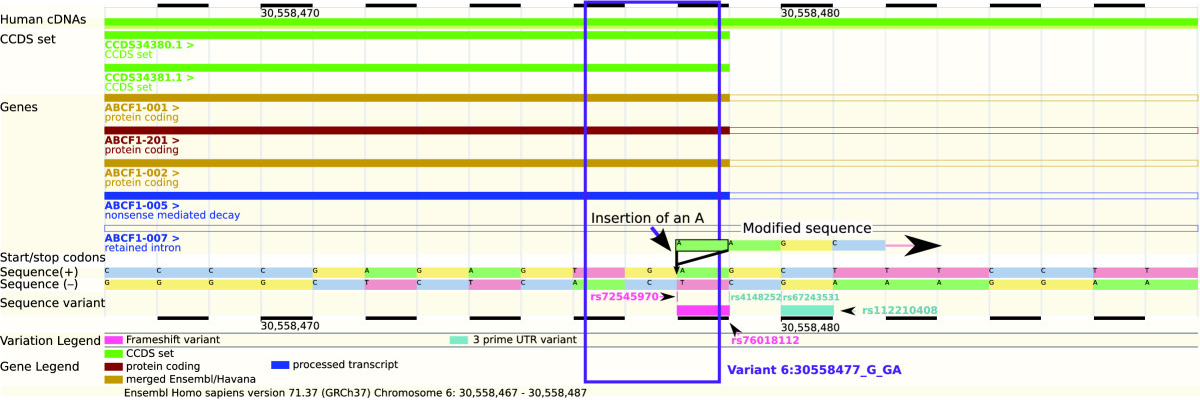
Variant annotation
- Variant annotations depend on quality of transcript annotations.
- Different annotation software may give different results.

Results
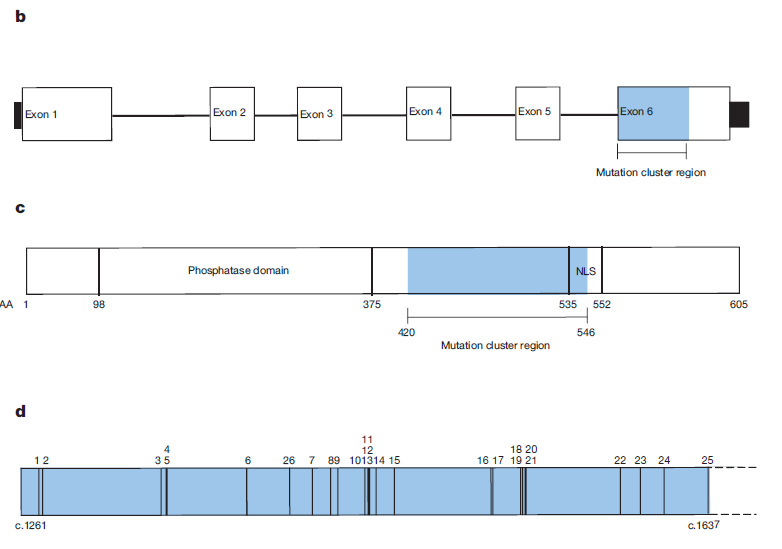
- Identified genes enriched for PTVs.
- Strongest PTV enrichment observed for PPM1D.
- Observed clustering of variants in final exon.
Sequenced affected exon in 7,781 cases and 5,861 controls
- 18 PTVs in 6,912 breast cancer cases
- 12 PTVs in 1,121 ovarian cancer cases
- 1 PTV in controls
PPM1D
- Induced in p53 dependent manner
- Contributes to deactivation of p53
- Part of negative feedback loop required to escape p53-dependent cell cycle arrest
- Truncated proteins show increased activity.
- PPM1D over-expression previously associated with breast cancer.

Non-coding variants: More information needed
Interpreting non-coding variants
- Impact of non-coding variants unclear
- Affected genes not obvious
- Effect on gene expression typically unknown
- Existing epigenetic data may help
- eQTL analyses can help to establish links between SNPs and genes


Using additional genome-wide data
ChIP-seq and RNA-seq data provide insight into the functional implications of genotypes. But need to consider
- Relevant cell type
- Relevant conditions

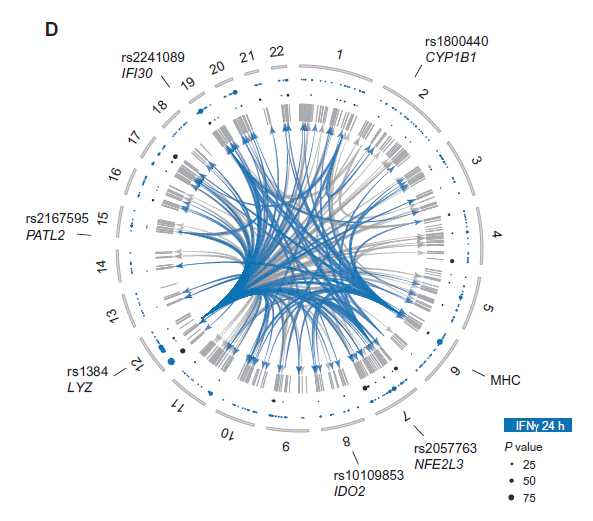
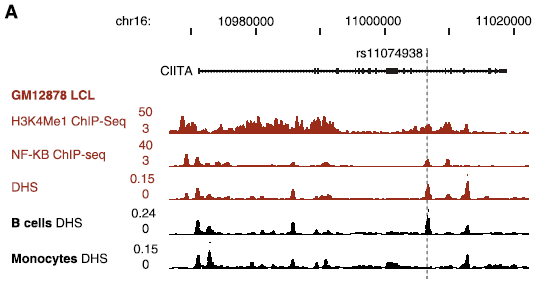
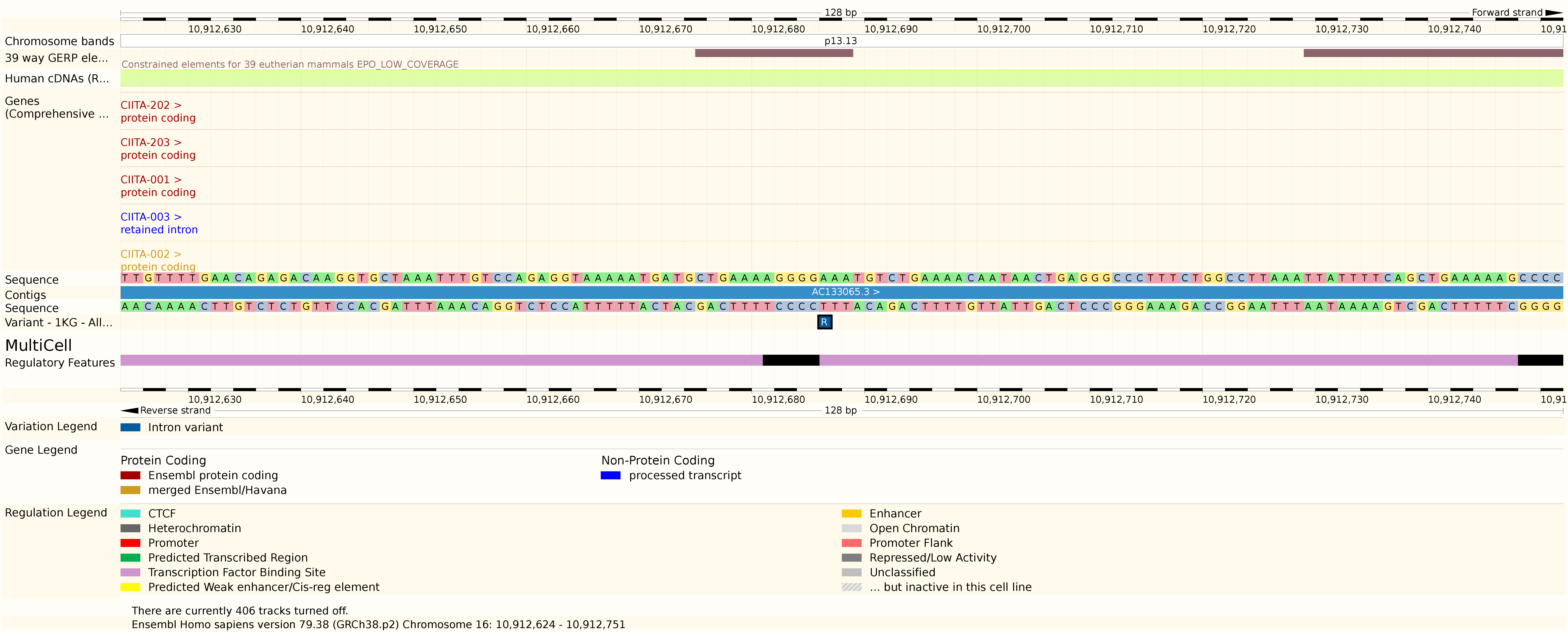
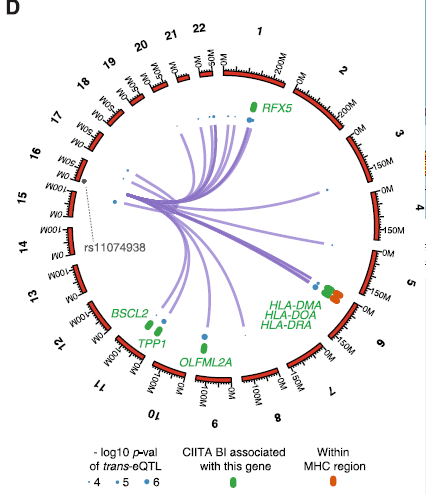
The role of rs11074938
- SNP (A/G) located in intron of CIITA
- Inside regulatory region
- ENCODE data shows DHS and NF-\(\kappa\)B binding
- CIITA is important regulator of MHC class II expression
- Could have implications for immune related diseases
(if this SNP affects CIITA)
Changes to gene expression

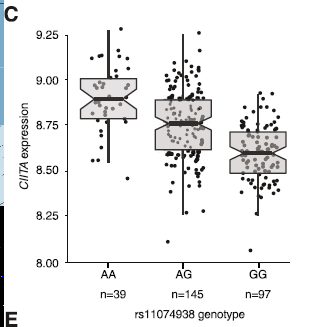
- eQTL analysis in B cells shows reduced expression of CIITA associated with G allele
- Evidence for changes in expression of CIITA target genes
- Reduced presence of MHC class II proteins on cell surface associated with G allele
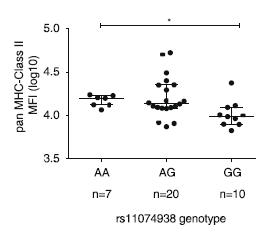
Transcription factor binding
- Reduced NF-\(\kappa\)B binding due to sequence change plausible
- Used existing ChIP-seq data\(^*\) to assess allele specific binding
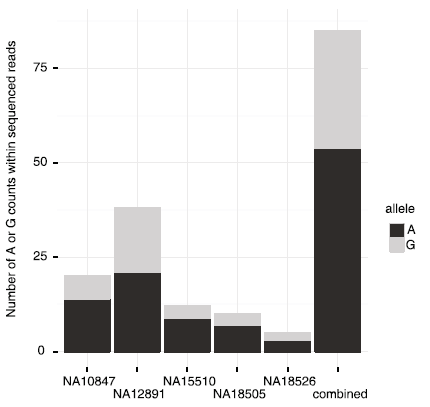
\(^*\) Kasowski et al., Science (2010). PMID: 20299548
Does it matter?
- No known GWAS associations
- Good to see that available data and methods can identify functional non-coding SNPs.
- Some evidence for association with the presence of nasal polyps in asthma patients\(^*\)
\(^*\) Bae et al., Mol Med Rep (2013). PMID: 23292525
Conclusions
This could be easier…
- Functional annotation of genomic variants is still difficult.
- Coding variants should be relatively easy, but annotations can be unreliable.
- Non-coding variants are still difficult.
- Understanding the effect of genomic variation requires a lot of work.
- Generating the data is easier than understanding it.
Challenges ahead
- Data volume is increasing, can the analysis keep pace?
- More and more public data sets available. Are we using them as much as we could?
- Better integration of all types of genomic data.
Acknowledgments
Breast cancer risk and variant annotation
 |
Peter Donnelly |  |
Nazneen Rahman |
 |
Manuel A. Rivas |  |
Katie Snape |
 |
Andrew Rimmer | Elise Ruark | |
 |
Davis McCarthy |
Acknowledgments
eQTL and CIITA
 |
Julian C. Knight |
 |
Daniel Wong |
| Wanseon Lee | |
 |
Benjamin P. Fairfax |
 |
Seiko Makino |
